Recombinant Severe acute respiratory syndrome coronavirus 2 Spike glycoprotein (S) (G476S), partial (Active)
In Stock-
貨號:CSB-MP3324GMY1(M4)
-
規格:¥1536
-
圖片:
-
(Tris-Glycine gel) Discontinuous SDS-PAGE (reduced) with 5% enrichment gel and 15% separation gel.
-
Activity
Measured by its binding ability in a functional ELISA. Immobilized SARS-CoV-2-S1-RBD (G476S) at 5 μg/ml can bind human ACE2 (CSB-MP866317HU), the EC50 is 47.11-92.3 ng/ml. Biological Activity Assay
-
-
其他:
產品詳情
-
純度:Greater than 85% as determined by SDS-PAGE.
-
內毒素:Less than 1.0 EU/ug as determined by LAL method.
-
生物活性:①Measured by its binding ability in a functional ELISA. Immobilized SARS-CoV-2-S1-RBD (G476S) at 5 μg/ml can bind human ACE2 (CSB-MP866317HU), the EC50 is 47.11-92.3 ng/ml.
-
基因名:
-
Uniprot No.:
-
別名:S; 2; Spike glycoprotein; S glycoprotein; E2; Peplomer protein)
-
種屬:Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) (SARS-CoV-2)
-
蛋白長度:Partial
-
來源:Mammalian cell
-
分子量:27.9 kDa
-
表達區域:319-541aa(G476S)
-
氨基酸序列RVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQASSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNF
-
蛋白標簽:C-terminal 10xHis-tagged
-
產品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
緩沖液:Lyophilized from a 0.2 μm sterile filtered 20 mM Tris-HCl, 0.5 M NaCl, 6% Trehalose, pH 8.0
-
復溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20°C/-80°C. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
儲存條件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保質期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
貨期:3-7 business days
-
注意事項:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet & COA:Please contact us to get it.
相關產品
靶點詳情
-
功能:attaches the virion to the cell membrane by interacting with host receptor, initiating the infection. Binding to human ACE2 receptor and internalization of the virus into the endosomes of the host cell induces conformational changes in the Spike glycoprotein. Binding to host NRP1 and NRP2 via C-terminal polybasic sequence enhances virion entry into host cell. This interaction may explain virus tropism of human olfactory epithelium cells, which express high level of NRP1 and NRP2 but low level of ACE2. The stalk domain of S contains three hinges, giving the head unexpected orientational freedom. Uses human TMPRSS2 for priming in human lung cells which is an essential step for viral entry. Can be alternatively processed by host furin. Proteolysis by cathepsin CTSL may unmask the fusion peptide of S2 and activate membranes fusion within endosomes.; mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.; Acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.; May down-regulate host tetherin (BST2) by lysosomal degradation, thereby counteracting its antiviral activity.
-
基因功能參考文獻:
- Study presents crystal structure of C-terminal domain of SARS-CoV-2 (SARS-CoV-2-CTD) spike S protein in complex with human ACE2 (hACE2); hACE2-binding mode similar overall to that observed for SARS-CoV. However, details at the binding interface show that key residue substitutions in SARS-CoV-2-CTD slightly strengthen the interaction and lead to higher affinity for receptor binding than SARS-CoV receptor-binding domain. PMID: 32378705
- crystal structure of the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 bound to the cell receptor ACE2 PMID: 32365751
- crystal structure of the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 (engineered to facilitate crystallization) in complex with ACE2 PMID: 32320687
- Out of the two isolates from India compared to the isolates from Wuhan, China, one was found to harbor a mutation in its receptor-binding domain (RBD) at position 407 where, arginine was replaced by isoleucine. This mutation has been seen to change the secondary structure of the protein at that region and this can potentially alter receptor binding of the virus. PMID: 32275855
- Structural modeling of the SARS-CoV-2 spike glycoprotein show similar receptor utilization between SARS-CoV-2 and SARS-CoV, despite a relatively low amino acid similarity in the receptor binding module. Compared to SARS-CoV and all other coronaviruses in Betacoronavirus lineage B, an extended structural loop containing basic amino acids were identified at the interface of the receptor binding (S1) and fusion (S2) domains. PMID: 32245784
- crystal structure of CR3022, a neutralizing antibody from a SARS patient, in complex with the receptor-binding domain of the SARS-CoV-2 spike (S) protein to 3.1 A; study provides insight into how SARS-CoV-2 can be targeted by the humoral immune response and revealed a conserved, but cryptic epitope shared between SARS-CoV-2 and SARS-CoV PMID: 32225176
- SARS-CoV and SARS-CoV-2 spike proteins have comparable binding affinities achieved by balancing energetics and dynamics. The SARS-CoV-2-ACE2 complex contains a higher number of contacts, a larger interface area, and decreased interface residue fluctuations relative to the SARS-CoV-ACE2 complex. PMID: 32225175
- Interaction interface between cat/dog/pangolin/Chinese hamster ACE2 and SARS-CoV/SARS-CoV-2 S protein was simulated through homology modeling. Authors identified that N82 of ACE2 showed closer contact with receptor-binding domain of S protein than human ACE2. PMID: 32221306
- SARS-CoV-2 S glycoprotein harbors a furin cleavage site at the boundary between the S1/S2 subunits, which is processed during biogenesis and sets this virus apart from SARS-CoV and SARS-related CoVs; determined cryo-EM structures of the SARS-CoV-2 S ectodomain trimer. PMID: 32201080
- Study demonstrates that SARS-CoV-2 uses the SARS-CoV receptor ACE2 for entry and the serine protease TMPRSS2 for S protein priming. PMID: 32155444
- The ACE2-B0AT1 complex exists as a dimer of heterodimers. Structural alignment of the RBD-ACE2-B0AT1 ternary complex with the S protein of SARS-CoV-2 suggests that two S protein trimers can simultaneously bind to an ACE2 homodimer. PMID: 32142651
- study demonstrated SARS-CoV-2 S protein entry on 293/hACE2 cells is mainly mediated through endocytosis, and PIKfyve, TPC2 and cathepsin L are critical for virus entry; found that SARS-CoV-2 S protein could trigger syncytia in 293/hACE2 cells independent of exogenous protease; there was limited cross-neutralization activity between convalescent sera from SARS and COVID-19 patients PMID: 32132184
- study determined a 3.5-angstrom-resolution cryo-electron microscopy structure of the 2019-nCoV S trimer in the prefusion conformation; provided biophysical and structural evidence that the 2019-nCoV S protein binds angiotensin-converting enzyme 2 (ACE2) with higher affinity than does severe acute respiratory syndrome (SARS)-CoV S PMID: 32075877
顯示更多
收起更多
-
亞細胞定位:Virion membrane; Single-pass type I membrane protein. Host endoplasmic reticulum-Golgi intermediate compartment membrane; Single-pass type I membrane protein. Host cell membrane; Single-pass type I membrane protein.
-
蛋白家族:Betacoronaviruses spike protein family
Most popular with customers
-
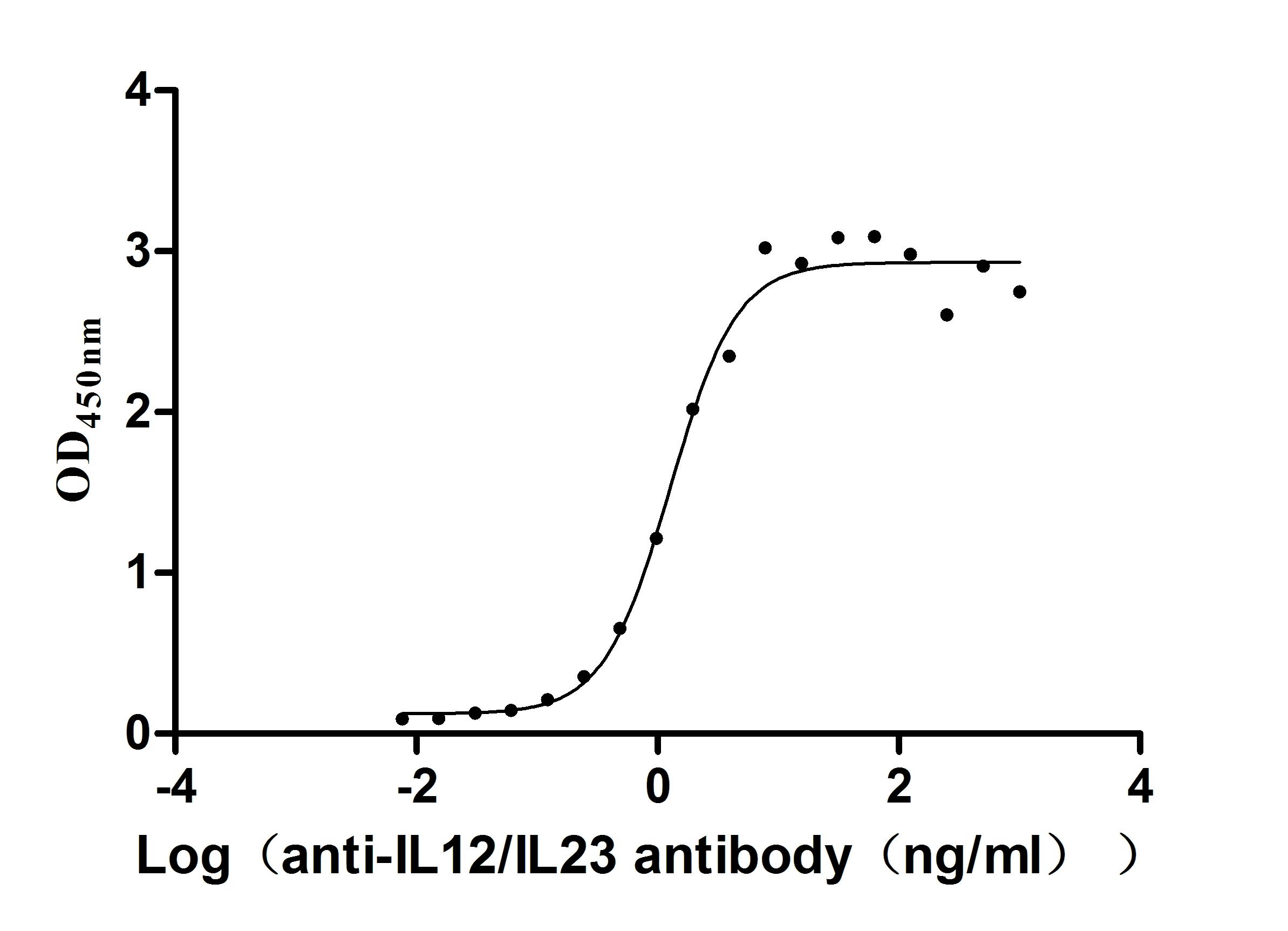
Recombinant Human IL12B&IL12A Heterodimer Protein (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
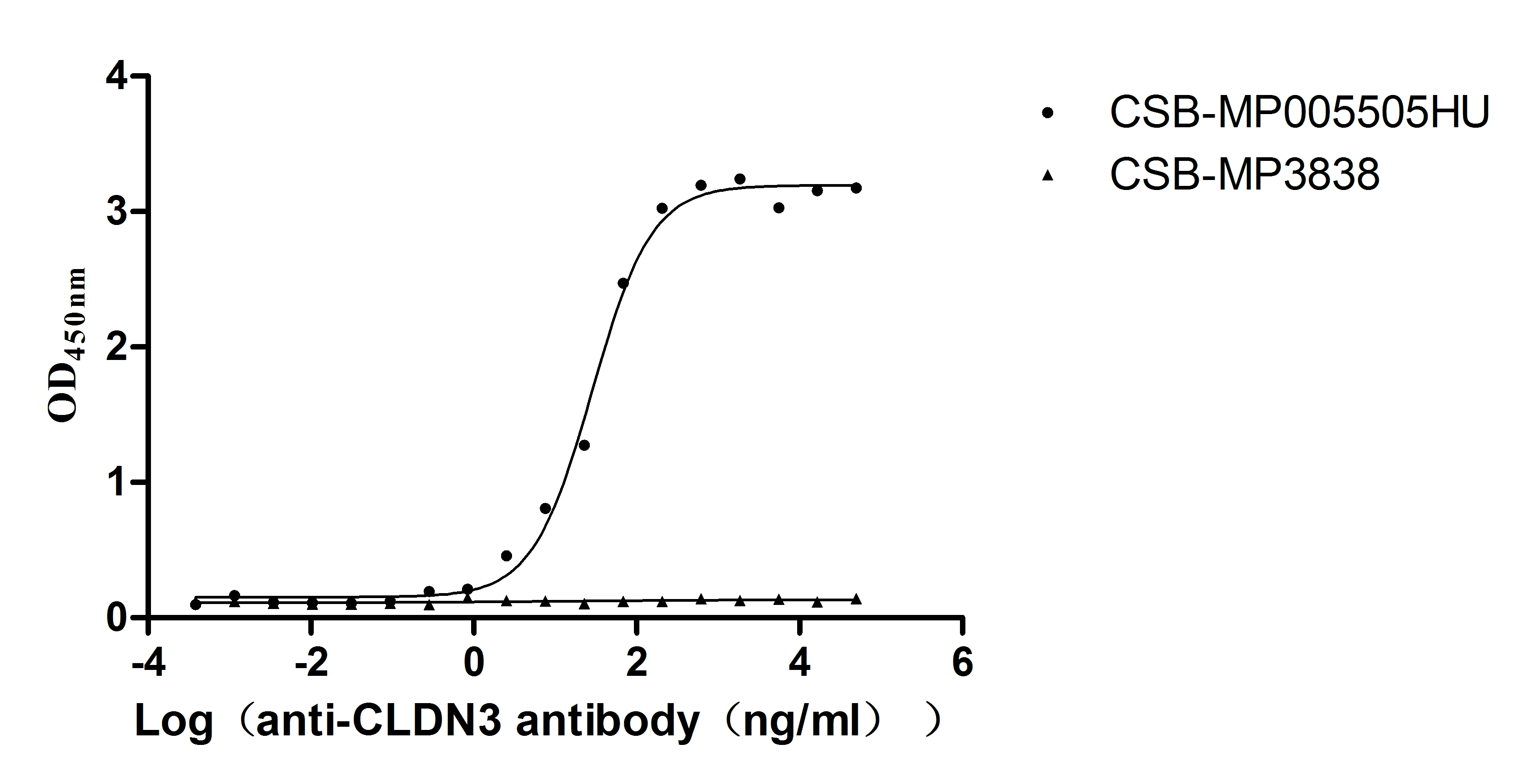
Recombinant Human Claudin-3 (CLDN3)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
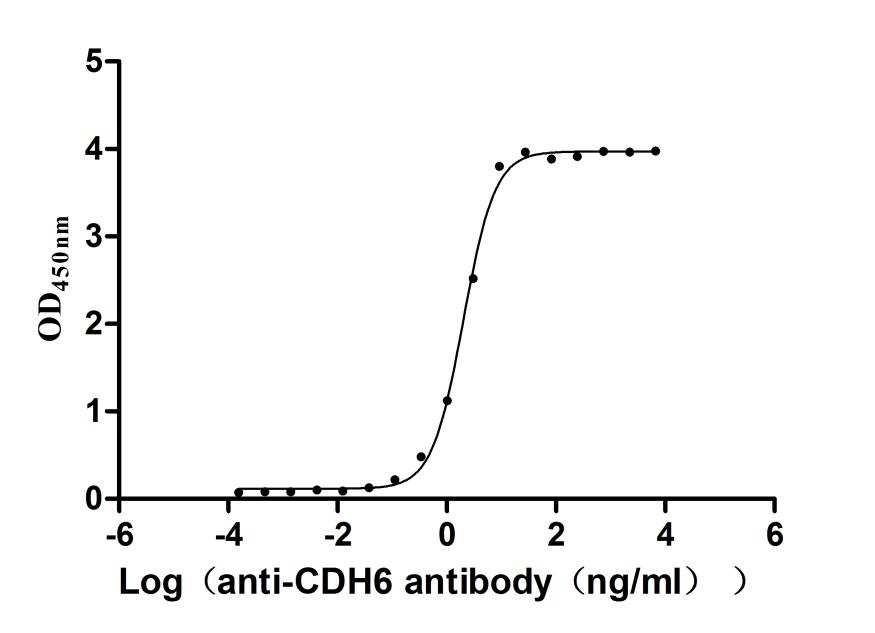
Recombinant Mouse Cadherin-6(Cdh6),partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)

-SDS.jpg)
-AC1.jpg)