Recombinant Saccharomyces cerevisiae Regulatory protein SIR3 (SIR3), partial
-
中文名稱:釀酒酵母SIR3重組蛋白
-
貨號:CSB-EP356918SVG-B
-
說明書:
-
規格:
-
來源:E.coli
-
共軛:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
產品詳情
-
純度:>85% (SDS-PAGE)
-
基因名:SIR3
-
Uniprot No.:
-
別名:SIR3; CMT1; MAR2; STE8; YLR442C; L9753.10Regulatory protein SIR3; Silent information regulator 3
-
種屬:Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
-
蛋白長度:Partial
-
蛋白標簽:Tag?type?will?be?determined?during?the?manufacturing?process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
產品提供形式:Lyophilized powder Warning: in_array() expects parameter 2 to be array, null given in /www/web/cusabio_cn/public_html/caches/caches_template/default/content/show_product_protein.php on line 662
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
復溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
儲存條件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保質期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
貨期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事項:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶點詳情
-
功能:The proteins SIR1 through SIR4 are required for transcriptional repression of the silent mating type loci, HML and HMR. The proteins SIR2 through SIR4 repress mulitple loci by modulating chromatin structure. Involves the compaction of chromatin fiber into a more condensed form.
-
基因功能參考文獻:
- Yeast Sir2 and Sir3 heterochromatin proteins directly modify the local chromatin environment of euchromatic DNA replication origins. PMID: 29795547
- Here, the authors analyze binding of the Sir proteins to reconstituted mono-, di-, tri-, and tetra-nucleosomal chromatin templates and show that key Sir-Sir interactions bridge only sites on different nucleosomes but not sites on the same nucleosome, and are therefore 'interrupted' with respect to sites on the same nucleosome. PMID: 27835568
- Repair of UV-induced DNA lesions in Saccharomyces cerevisiae telomeres is moderated by Sir2 and Sir3, and inhibited by yKu-Sir4 interaction. PMID: 28334768
- silencing requires Sir2, a highly-conserved NAD(+)-dependent histone deacetylase. At locations other than the rDNA, silencing also requires additional Sir proteins, Sir1, Sir3, and Sir4 that together form a repressive heterochromatin-like structure termed silent chromatin. PMID: 27516616
- results suggest that Sir3 exhibits novel binding parameters and gene regulatory functions at the CN binding domains. PMID: 26957548
- Sir3 interacts with nucleosomal arrays with a stoichiometry of two Sir3 monomers per nucleosome. We also find that Sir3 fibres are less compact than canonical magnesium-induced 30 nm fibres. PMID: 25163529
- Direct interactions promote eviction of the Sir3 heterochromatin protein by the SWI/SNF chromatin remodeling enzyme. PMID: 25453095
- Telomere clusters, their dynamics, and their nuclear distribution result from random motion, aggregation, and dissociation of telomeric regions, specifically determined by the amount of Sir3. PMID: 23576549
- The crystal structure of the N-terminally acetylated BAH domain of Sir3 bound to the nucleosome core particle reveals that the N-terminal acetylation stabilizes the interaction of Sir3 with the nucleosome. PMID: 23934150
- By X-ray crystallography, we show that the acetylated N terminus of Sir3 does not interact with the nucleosome directly. Instead, it stabilizes a nucleosome-binding loop in the BAH domain. PMID: 23934152
- Sir3 binding generates a more stable nucleosome by clamping H4R17 and R19 to nucleosomal DNA, and raise the possibility that such induced changes in histone-DNA contacts play major roles in the regulation of chromatin structure. PMID: 23650358
- Dimerization of Sir3 via its C-terminal winged helix domain is essential for yeast heterochromatin formation. PMID: 23299941
- examined gene expression and molecular markers of silencing at the silent mating type loci under conditions of limiting Sir3 protein PMID: 22586263
- crystal structure of complex of Sir3 BAH domain and nucleosome core particle at 3.0 A resolution; structure explains how covalent modifications on H4K16 and H3K79 regulate formation of silencing complex containing the nucleosome as a central component PMID: 22096199
- Sir3 promotes telomere clustering independently of silencing in yeast PMID: 21300849
- Genome-wide nucleosome mapping revealed that Sir3 binding to subtelomeric regions was associated with overpackaging of subtelomeric promoters. PMID: 21336256
- Data suggest that histone H3 Lys-4 methylation prevents Sir3p association at euchromatic sites and therefore concentrates Sir3p at unmodified, heterochromatic regions of the genome. PMID: 15280381
- sir2 and sir4, but not sir3, have roles in silencing of DNA breakage and recombination PMID: 15647382
- The structure and function of the Sir3 BAH domain have been established. PMID: 16581798
- These findings imply that the Sir3p-BAH domain structure has evolved for functions distinct from those of the Orc1p-BAH domain. PMID: 16641491
- Sir3-Sir4 complexes form progressively higher order assemblies with increasing protein concentration, with implications for the mechanism of gene silencing. PMID: 16717101
- Chromatin immunoprecipitation analyses of wild-type and ctd-Y964A mutant cells indicate an association of the C-terminal domain with the deacetylated histone tails of H3 and H4 that is necessary for the recruitment of Sir3 PMID: 16908543
- Analysis of full-length recombinant Sir3p domain organization and quaternary structure supports a nucleosome bridging model for Sir3p-dependent regulation of chromatin architecture. PMID: 17176117
- Results show that the BAH domain of Sir3 is a nucleosome- and histone-tail-binding domain and its binding to nucleosomes is regulated by the N terminus residues of histone H4 and the globular domain of histone H3 on the exposed surface of the nucleosome. PMID: 18158899
- These results define how SIR3p may function as a chromatin architectural protein and provide new insight into the interplay between endogenous and protein-mediated chromatin fiber condensation pathways. PMID: 18362167
- BAH domain of Sir3 binds to histone H3K79; acetylation of the BAH domain is required for the binding specificity of Sir3 for nucleosomes unmethylated at H3K79. PMID: 18391024
- Results underscore the importance of proper interactions between Sir3 and the nucleosome in silent chromatin assembly. PMID: 18794362
- Sir3p BAH domain directly binds the nucleosomal LRS domain. PMID: 19079580
- Role of nucleic acid binding in Sir3-dependent interactions with chromatin fibers is reported. PMID: 19099415
- The functional specialization of Sir3,as a silencing protein was facilitated by the tandem duplication of the OIR domain in the Sir1 family, allowing distinct Sir1-Sir3 and Sir1-Orc1 interactions through OIR-BAH domain interactions. PMID: 19171939
- These results, together with the previously characterized interaction between the C-terminal region of Sir3 and the histone H3/H4 tails, suggest that Sir3 utilizes multiple domains to interact with nucleosomes. PMID: 19273586
顯示更多
收起更多
-
亞細胞定位:Nucleus.
-
數據庫鏈接:
KEGG: sce:YLR442C
STRING: 4932.YLR442C
Most popular with customers
-
Recombinant Human Heat-stable enterotoxin receptor (GUCY2C), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Tyrosine-protein kinase Mer (MERTK), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Poliovirus receptor (PVR) (I340M), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human HLA class II histocompatibility antigen gamma chain (CD74), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
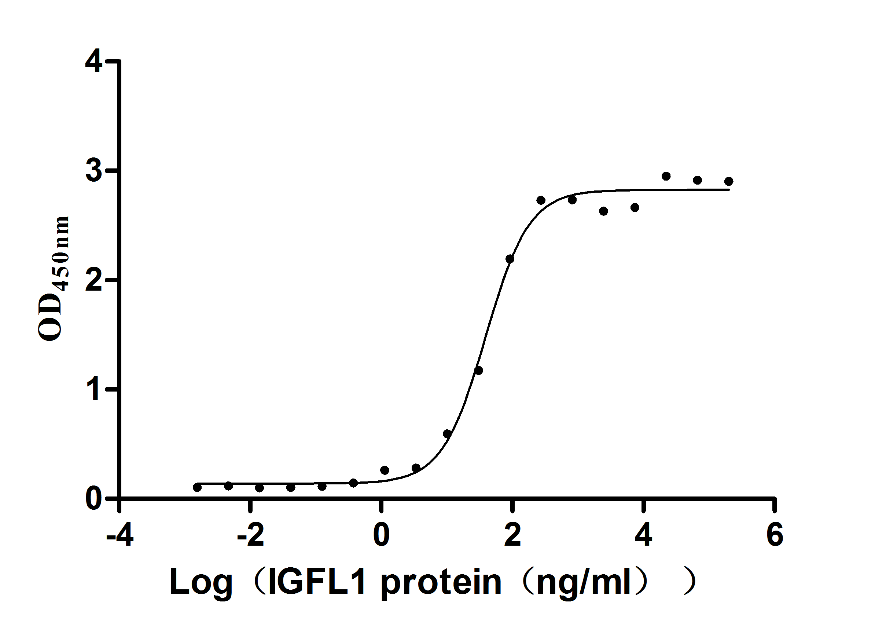
Recombinant Human IGF-like family receptor 1 (IGFLR1), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
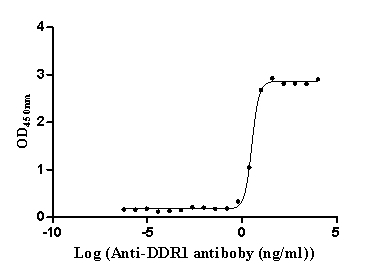
Recombinant Human Epithelial discoidin domain-containing receptor 1 (DDR1), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
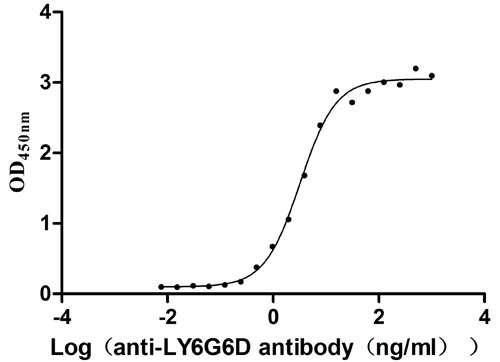
Recombinant Human Lymphocyte antigen 6 complex locus protein G6d (LY6G6D) (Active)
Express system: Yeast
Species: Homo sapiens (Human)
-
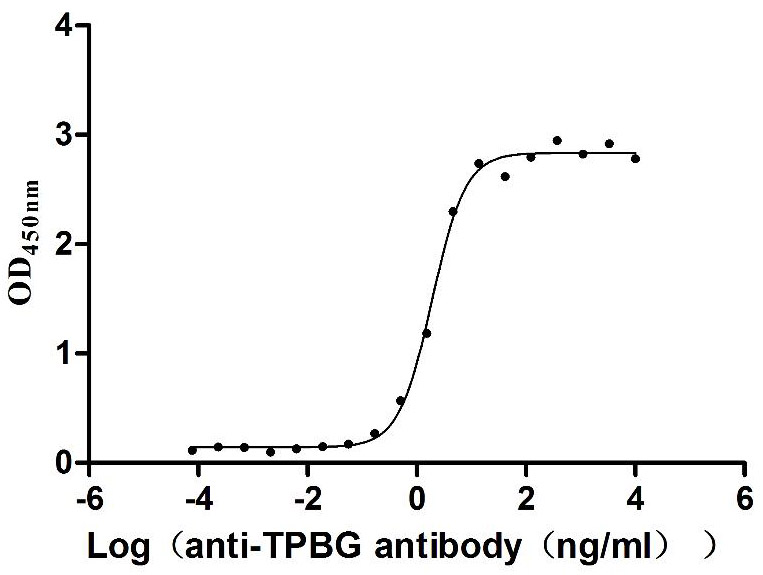
Recombinant Macaca fascicularis Trophoblast glycoprotein (TPBG), partial (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)


-AC1.jpg)
-AC1.jpg)