PIAS1 Recombinant Monoclonal Antibody
-
中文名稱:PIAS1 Recombinant Monoclonal Antibody
-
貨號:CSB-RA106449A0HU
-
規格:¥1320
-
圖片:
-
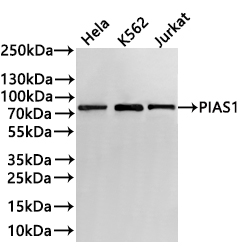
Western Blot
Positive WB detected in: Hela whole cell lysate(30μg), K562 whole cell lysate(30μg), Jurkat whole cell lysate(30μg)
All lanes: PIAS1 antibody at 1:1000
Secondary
Goat polyclonal to rabbit IgG at 1/40000 dilution
Predicted band size: 72 kDa
Observed band size: 75 kDa
Exposure time:3min -
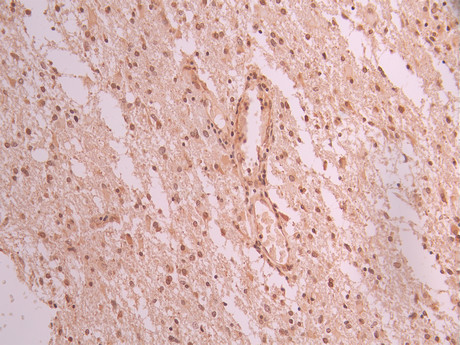
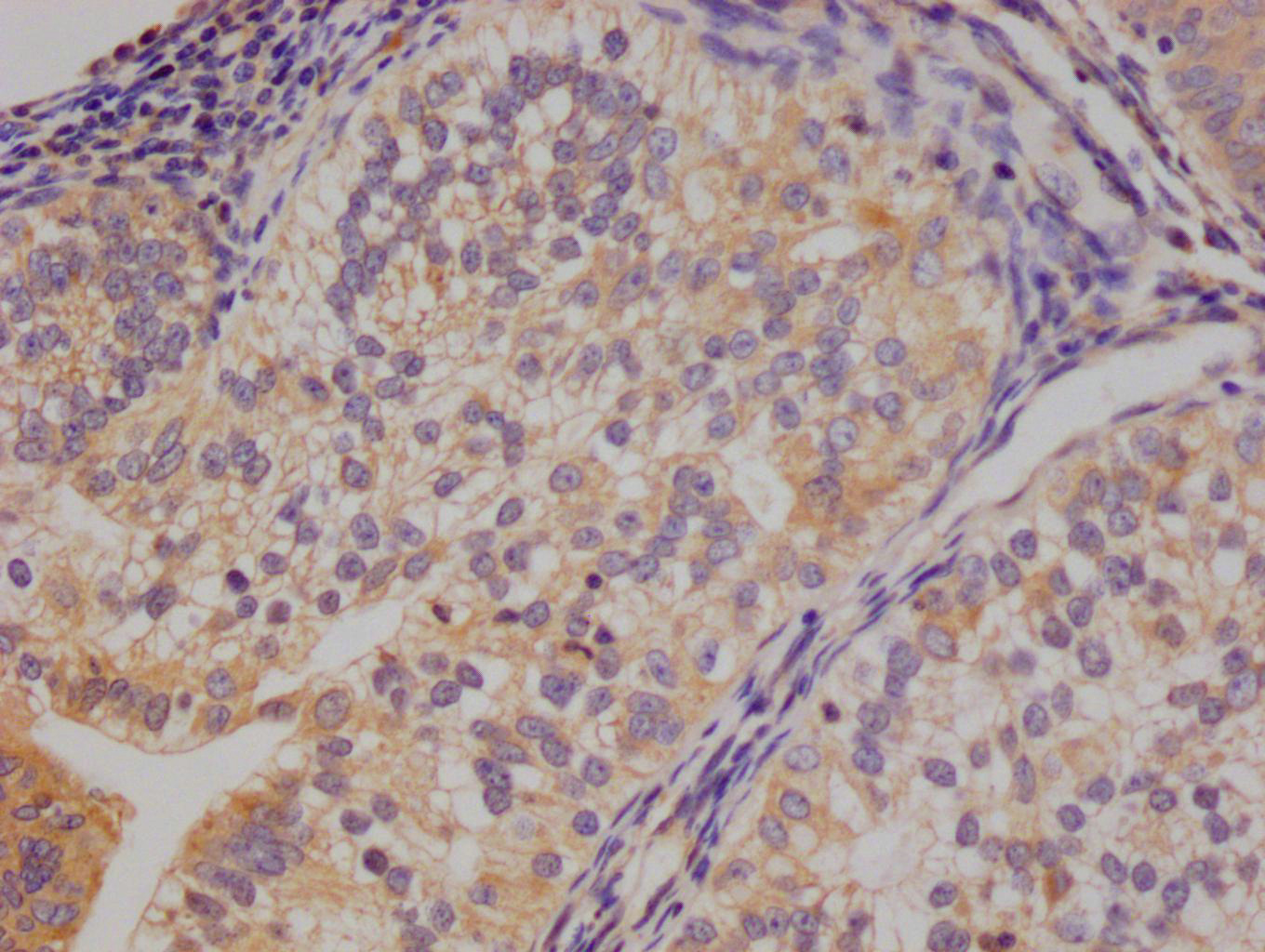
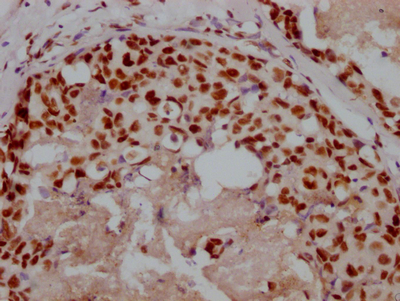
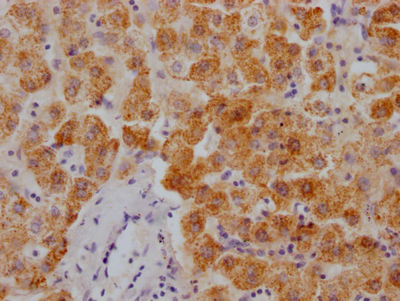
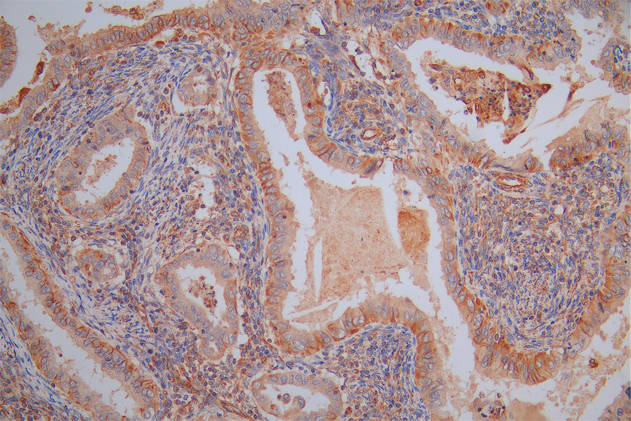
IHC image of CSB-RA106449A0HU diluted at 1:100 and staining in paraffin-embedded human glioma cancer performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a Goat anti-rabbit polymer IgG labeled by HRP and visualized using 0.05% DAB.
-
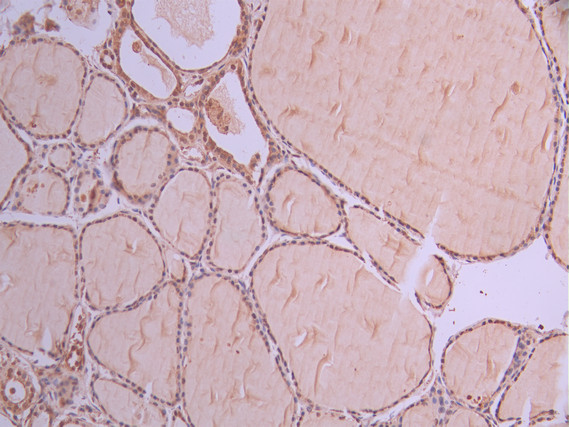
IHC image of CSB-RA106449A0HU diluted at 1:100 and staining in paraffin-embedded human thyroid tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a Goat anti-rabbit polymer IgG labeled by HRP and visualized using 0.05% DAB.
-
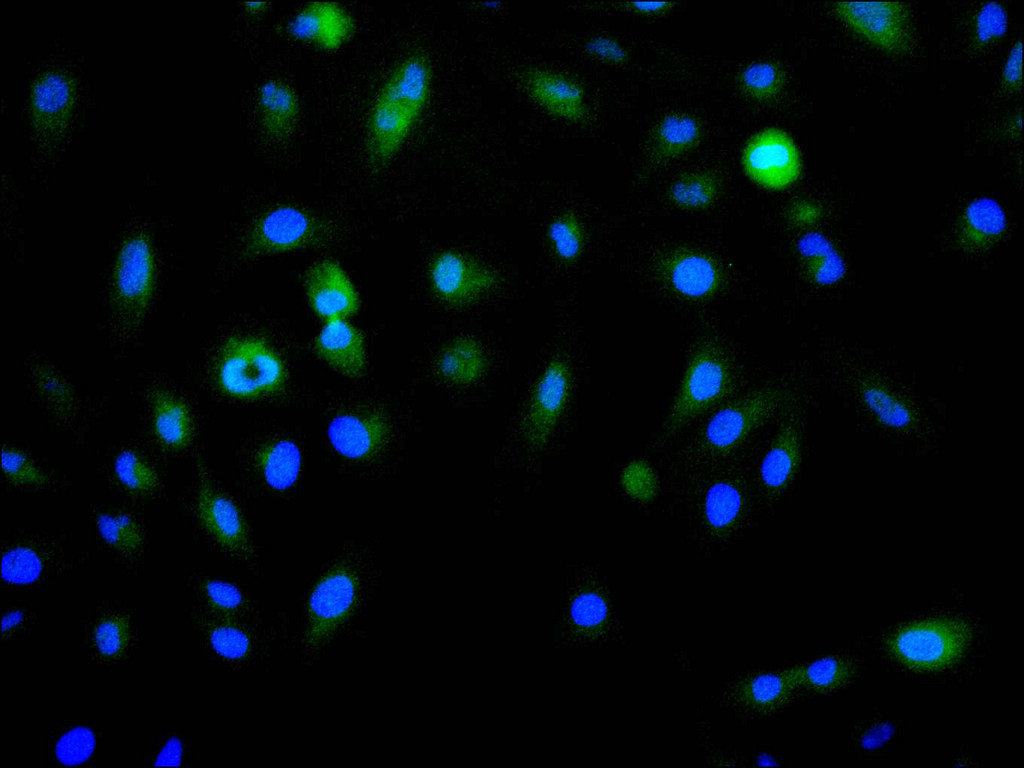
Immunofluorescence staining of A549 cell with CSB-RA106449A0HU at 1:50, counter-stained with DAPI. The cells were fixed in 4% formaldehyde, permeabilized using 0.2% Triton X-100 and blocked in 10% normal Goat Serum. The cells were then incubated with the antibody overnight at 4°C. The secondary antibody was Alexa Fluor 488-congugated AffiniPure Goat Anti-Rabbit IgG(H+L).
-
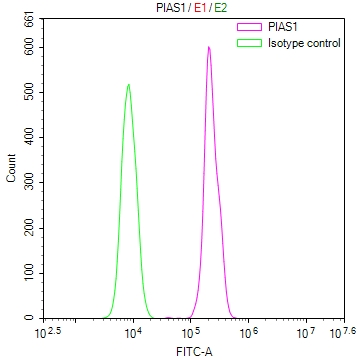
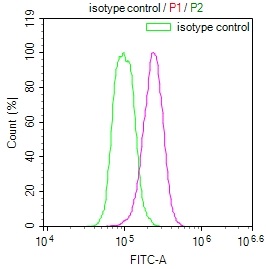
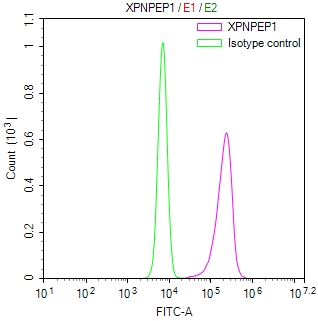
Overlay Peak curve showing Hela cells stained with CSB-RA106449A0HU (red line) at 1:100. The cells were fixed in 4% formaldehyde and permeated by 0.2% TritonX-100 for?10min. Then 10% normal goat serum to block non-specific protein-protein interactions followed by the antibody (1ug/1*106cells) for 45min at 4℃. The secondary antibody used was FITC-conjugated goat anti-rabbit IgG (H+L) at 1/200 dilution for 35min at 4℃.Control antibody (green line) was Rabbit IgG (1ug/1*106cells) used under the same conditions. Acquisition of >10,000 events was performed.
-
-
其他:
產品詳情
-
Uniprot No.:
-
基因名:
-
別名:E3 SUMO-protein ligase PIAS1 (EC 2.3.2.-) (DEAD/H box-binding protein 1) (E3 SUMO-protein transferase PIAS1) (Gu-binding protein) (GBP) (Protein inhibitor of activated STAT protein 1) (RNA helicase II-binding protein), PIAS1, DDXBP1
-
反應種屬:Human
-
免疫原:A synthesized peptide from human PIAS1 protein
-
免疫原種屬:Homo sapiens (Human)
-
標記方式:Non-conjugated
-
克隆類型:Monoclonal
-
抗體亞型:Rabbit IgG
-
純化方式:Affinity-chromatography
-
克隆號:9B11
-
濃度:It differs from different batches. Please contact us to confirm it.
-
保存緩沖液:Preservative: 0.03% Proclin 300
Constituents: 50% Glycerol, 0.01M PBS, PH 7.4 -
產品提供形式:Liquid
-
應用范圍:ELISA, WB, IHC, IF, FC
-
推薦稀釋比:
Application Recommended Dilution WB 1:500-1:2000 IHC 1:50-1:200 IF 1:50-1:200 FC 1:50-1:200 -
Protocols:
-
儲存條件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
貨期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
-
用途:For Research Use Only. Not for use in diagnostic or therapeutic procedures.
相關產品
靶點詳情
-
功能:Functions as an E3-type small ubiquitin-like modifier (SUMO) ligase, stabilizing the interaction between UBE2I and the substrate, and as a SUMO-tethering factor. Plays a crucial role as a transcriptional coregulation in various cellular pathways, including the STAT pathway, the p53 pathway and the steroid hormone signaling pathway. In vitro, binds A/T-rich DNA. The effects of this transcriptional coregulation, transactivation or silencing, may vary depending upon the biological context. Sumoylates PML (at'Lys-65' and 'Lys-160') and PML-RAR and promotes their ubiquitin-mediated degradation. PIAS1-mediated sumoylation of PML promotes its interaction with CSNK2A1/CK2 which in turn promotes PML phosphorylation and degradation. Enhances the sumoylation of MTA1 and may participate in its paralog-selective sumoylation. Plays a dynamic role in adipogenesis by promoting the SUMOylation and degradation of CEBPB.
-
基因功能參考文獻:
- Rad18, independently of its ubiquitin ligase activity, promotes DNA polymerase eta SUMOylation by facilitating its interaction with its SUMO ligase PIAS1 and is required for DNA polymerase eta function at difficult to replicate loci. PMID: 27811911
- PIAS1 as a key regulator of Epstein-Barr Virus lytic replication. PIAS1 acts as an inhibitor for transcription factors involved in lytic gene expression. PMID: 29262325
- these results indicate that PIAS1 is a positive regulator of MYC. PMID: 27239040
- PIAS1 is a prognostic biomarker in breast cancer PMID: 28493978
- PIAS1 is a determinant of poor survival and acts as a positive feedback regulator of AR signaling through enhanced AR stabilization in prostate cancer PMID: 26257066
- PIAS1 overexpression exacerbated mutant Huntingtin-associated phenotypes and aberrant protein accumulation PMID: 27146268
- HBs protein-induced hPIAS1 transcription requires TAL1, E47, MYOG, NFI, and MAPK signal pathways PMID: 27276529
- identified the SUMO ligase PIAS1 as a constituent PML-NB antiviral protein. This finding distinguishes a SUMO ligase that may mediate signaling events important in promyelocytic leukemia nuclear body mediated intrinsic immunity. PMID: 27099310
- PIAS1 enhances p300 recruitment to c-Myb-bound sites through interaction with both proteins. In addition, the E3 activity of PIAS1 enhances further its coactivation PMID: 27032383
- Results show that apocrine breast cancer and prostate cancer cells share a core AR cistrome and target gene signature linked to cancer cell growth, and PIAS1 plays a similar coregulatory role for AR in both cancer cell types. PMID: 26219822
- c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4 PMID: 25895136
- Data demonstrate that PIAS1 interacts with TRF2 and mediates its sumoylation serving as a molecular switch that controls the level of TRF2 at telomeres. PMID: 26450775
- the expression of SENP8, SAE1, PIAS1, PIAS2 and ZMIZ1 is deregulated in the majority of PTC tissues, likely contributing to the PTC phenotype. PMID: 26403403
- Our data suggest that necdin suppresses PIAS1 both by inhibiting SUMO E3 ligase activity and by promoting ubiquitin-dependent degradation. PMID: 24911587
- the presented data indicate that PIAS1 is crucial for parental and docetaxel resistant prostate cancer cell survival PMID: 25474038
- Elevated PIAS1 expression was observed in breast tumor samples. PMID: 24586797
- PIAS1 is the E3 ligase responsible for SUMOylation of HMGN2. PMID: 24872413
- Further study indicated that PIAS1 interacted with IRF3 and inhibited the DNA binding activity of IRF3. PMID: 24036127
- Levels of STAT1 andor the protein expression of its negative regulators, PIAS1 and SOCS3, may be a good predictor of hepatitis C virus response to therapy PMID: 23472246
- Smad2 and PIAS1 proteins were significantly upregulated resulting in dramatically increased interactions between Smad2/4 and PIAS1 in the presence of zinc. PMID: 24052079
- Phosphorylation of serine residues adjacent to the PIAS1 SUMO-interacting motif favors formation of the non covalent PIAS1.SUMO.UBC9 ternary complex PMID: 24174529
- Data suggest that the pro-apoptotic protein Daxx specifically interacts with one or more substrates SUMOylated by PIAS1 and this interaction leads to apoptosis following UV irradiation. PMID: 22976298
- data suggest that PIAS1 may function as a tumor suppressor to regulate gastric cancer cell metastasis by targeting the MAPK signaling pathway PMID: 22972521
- MAPK-activated protein kinase-2 limits endothelial inflammation via the PIAS1 S522 phosphorylation-mediated increase in PIAS1 transrepression and SUMO ligase activity. PMID: 23202365
- PIAS1 is a SUMO ligase for GATA4 that differentially regulates GATA4 transcriptional activity independent of SUMO ligase activity and GATA4 sumoylation. PMID: 22539995
- The data reveal an important new role for PIAS1 in the regulation of cell proliferation in prostate cancer. PMID: 22449952
- PIAS1 negatively regulates ubiquitination of Msx1 homeoprotein independent of its SUMO ligase activity. PMID: 21717107
- There are differences in the PIAS3 expression from different stages of gastric precancerous conditions/lesions to GC, which may reveal a close relationship between expression reduction or loss of PIAS3 and gastric tumorigenesis. PMID: 21925039
- PIAS1 determines the level of JNK activity in human endometrial stromal cells , couples ROS signaling to the SUMO pathway, and promotes oxidative cell death. PMID: 21676946
- PIAS1 is a common partner for two cancer-related nuclear factors, c-Myb and FLASH. PMID: 21338522
- Data show that regulation of SATB1 sumoylation and caspase cleavage is controlled by SATB1 phosphorylation; specifically, PIAS1 interaction with SATB1 is inhibited by phosphorylation. PMID: 20351170
- protein inhibitor of activated Stat1 (PIAS1) interacts with the tetramerization and C-terminal regulatory domains of p53 in yeast two-hybrid analyses PMID: 11788578
- Protein inhibitors of activated STAT resemble scaffold attachment factors and function as interacting nuclear receptor coregulators. PMID: 11877418
- PIAS1 and PIASxalpha modulate the AR-dependent transactivation, which, at least in part, can be attributed to their SUMO-E3 activity toward AR. PMID: 12177000
- PIAS1 has a role in sumoylation of MDM2 in the cell nucleus PMID: 12393906
- found to strongly stimulate sumoylation of STAT1 at Lys703; results suggest a negative regulatory function for sumoylation. PMID: 12855578
- PIAS1 interacts with the N-terminal domain of human mineralocorticoid receptor and represses its ligand-dependent transcription. PMID: 14500761
- three-dimensional structure and its binding duality are discussed in conjunction with the biological functions of PIAS1 as a SUMO ligase PMID: 15133049
- PIAS1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. PMID: 15572666
- PIAS1 interacts with DNA cross-link repair SNM1A in nuclear focus formation. PMID: 15572677
- Results suggest that recombinant human interleukin-12 upregulates STAT-1 expression and that increased expression may be dose dependent. PMID: 15901746
- PIAS1 modulates transcriptional activation of smooth muscle cells marker genes through cooperative interactions with both serum response factor and class I basic helix-loop-helix proteins proteins. PMID: 16135793
- Pias1 binds to and sumoylates metabotropic glutamate receptor 8 PMID: 16144832
- In this study, we demonstrate that MEF2A undergoes sumoylation primarily at a single lysine residue (K395) both in vitro and in vivo. We also show that the nuclear E3 ligase, PIAS1, promotes sumoylation of MEF2A. PMID: 16563226
- PIAS1 is required for the appropriate localization and retention of Msx1 at the nuclear periphery in myoblast cells. PMID: 16600910
- TGF-beta rapidly suppresses IFN-gamma-driven STAT1 signaling by reducing DNA binding via promotion of STAT1--PIAS1 interactions and not inhibition of STAT1 activation. PMID: 17371985
- PIASy cooperates with PIAS1 to down-regulate the specificity and magnitude of NF-kappa B/STAT1-mediated gene activation. PMID: 17606919
- The data show that HCV NS3/4a is able to block the Jak-Stat signaling pathway at the stage of Stat-1 serine 727 phosphorylation. PMID: 18190974
- PIAS1 staining of the colon cancer tissue microarrays indicated a strong correlation of normal colon cells, and adenomas, with high expression of both PIAS1 and IRF-1 PMID: 19288270
- novel function of PIAS1 in the induction of JNK-dependent apoptosis, independent of the previously known inhibitory activity of PIAS1 in STAT-mediated gene activation. PMID: 11451946
顯示更多
收起更多
-
亞細胞定位:Nucleus speckle. Nucleus, PML body. Note=Interaction with CSRP2 may induce a partial redistribution along the cytoskeleton.
-
蛋白家族:PIAS family
-
組織特異性:Expressed in numerous tissues with highest level in testis.
-
數據庫鏈接:
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC, IF, FC
Species Reactivity: Human, Mouse, Rat
-
-
-
-
-
-