Acetyl-HIST1H3A(K18) Recombinant Monoclonal Antibody
-
中文名稱:Acetyl-HIST1H3A(K18) 重組抗體
-
貨號:CSB-RA547773A0HU
-
規格:¥1320
-
圖片:
-
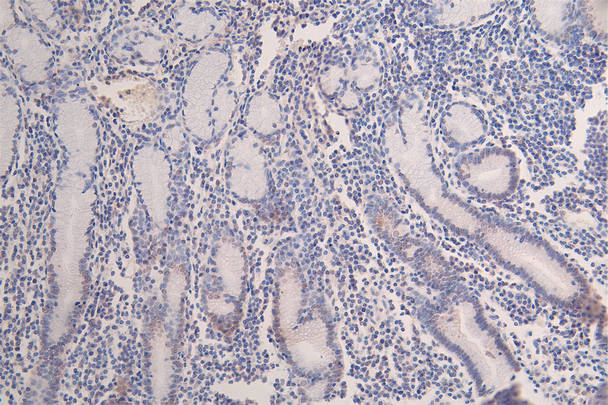
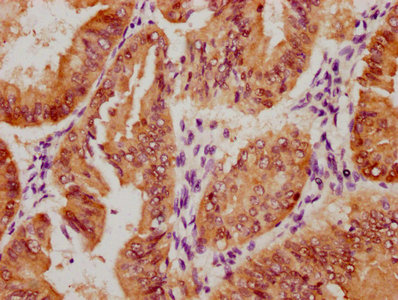
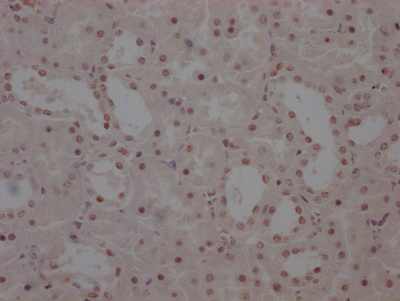
IHC image of CSB-RA547773A0HU diluted at 1:100 and staining in paraffin-embedded human breast cancer performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a Goat anti-rabbit polymer IgG labeled by HRP and visualized using 0.07% DAB.
-
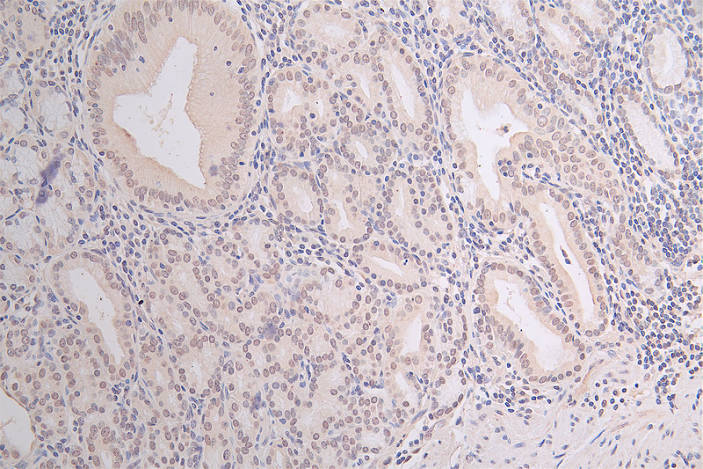
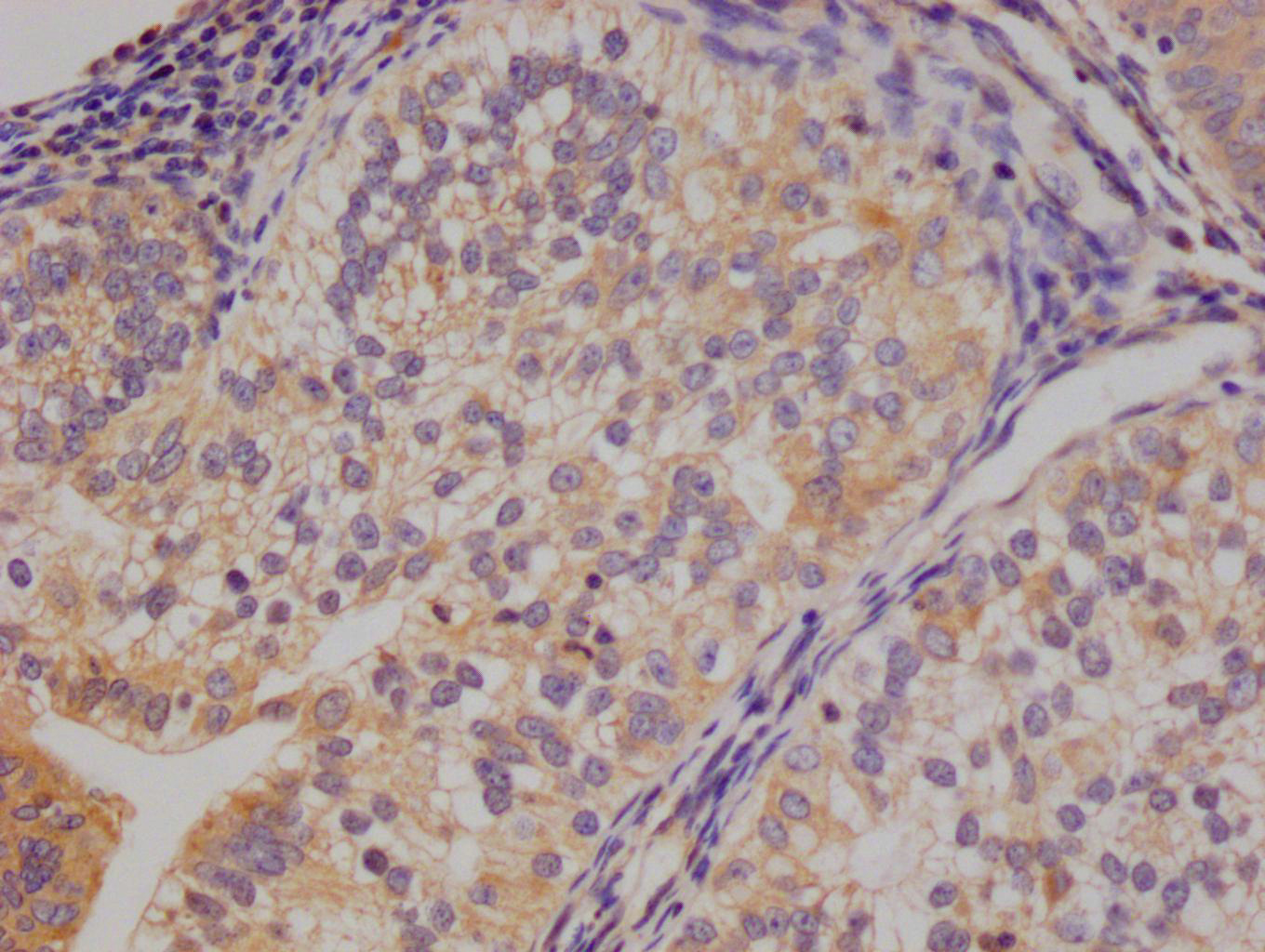
IHC image of CSB-RA547773A0HU diluted at 1:100 and staining in paraffin-embedded human gastric cancer performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a Goat anti-rabbit polymer IgG labeled by HRP and visualized using 0.07% DAB.
-
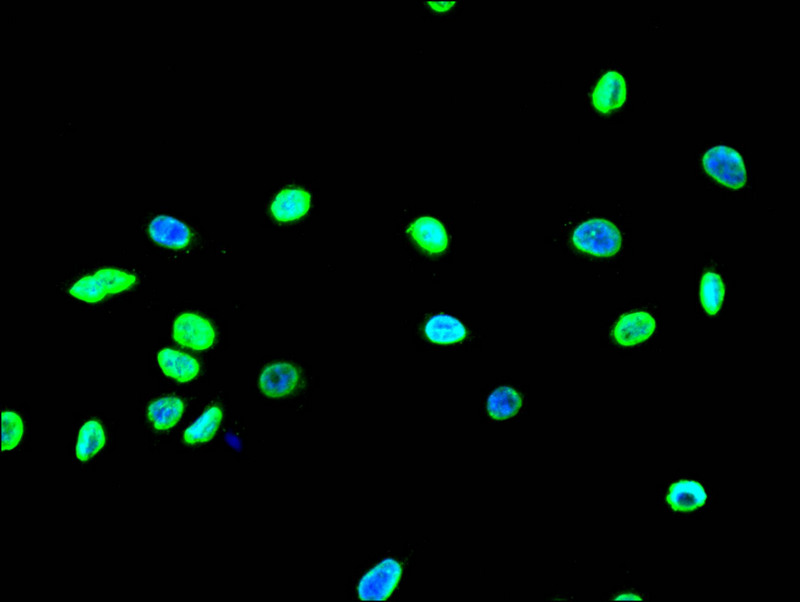
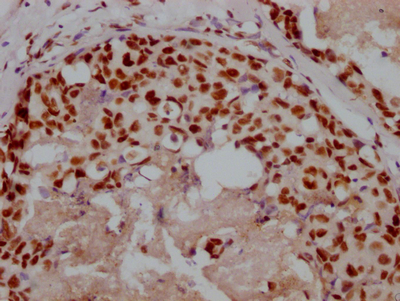
Immunofluorescence staining of A549 with CSB-RA547773A0HU at 1:10, counter-stained with DAPI. The cells were fixed in 4% formaldehyde and blocked in 10% normal Goat Serum. The cells were then incubated with the antibody overnight at 4°C. The secondary antibody was Alexa Fluor 490-congugated AffiniPure Goat Anti-Rabbit IgG(H+L).
-
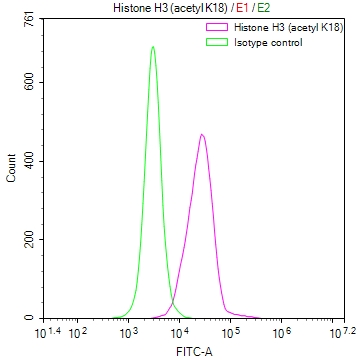
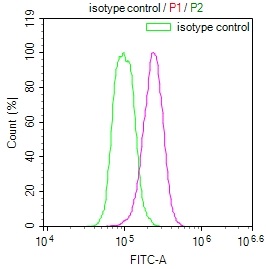
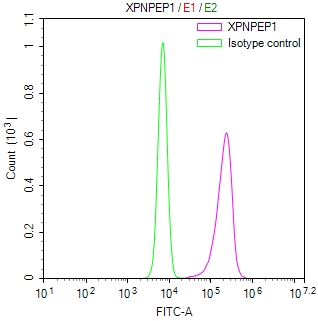
Overlay Peak curve showing A549 cells stained with CSB-RA547773A0HU (red line) at 1:50. The cells were fixed in 4% formaldehyde and permeated by 0.2% TritonX-100. Then 10% normal goat serum to block non-specific protein-protein interactions followed by the antibody (1μg/1*106cells) for 45min at 4℃. The secondary antibody used was FITC-conjugated Goat Anti-rabbit IgG(H+L) at 1:200 dilution for 35min at 4℃.Control antibody (green line) was rabbit IgG (1μg/1*106cells) used under the same conditions. Acquisition of >10,000 events was performed.
-
-
其他:
產品詳情
-
產品描述:CSB-RA547773A0HU Acetyl-HIST1H3A重組單克隆抗體是一款針對組蛋白H3第1亞型乙酰化修飾位點開發的高特異性科研工具,適用于表觀遺傳調控機制研究。該抗體靶向HIST1H3A蛋白的乙酰化修飾形式,該修飾通過改變染色質結構參與基因轉錄激活,在細胞分化、周期調控及腫瘤發生等生物學過程中發揮重要作用。經嚴格驗證,該抗體在多種實驗場景中表現優異,推薦使用稀釋比例為:免疫組化(IHC)1:50-1:200、免疫熒光(IF)1:50-1:200、流式細胞術(FC)1:50-1:200,同時支持ELISA檢測。其重組單克隆特性確保了高批次間一致性,適用于染色質動態分析、表觀遺傳標記檢測、癌癥相關基因表達調控研究等領域,可廣泛應用于細胞模型或組織樣本中乙酰化H3蛋白的定位與定量分析。該產品為探索組蛋白修飾介導的基因沉默/激活機制、腫瘤表觀遺傳異常等研究提供可靠工具,滿足分子生物學和細胞生物學領域對高特異性表觀遺傳標記物的研究需求。
-
Uniprot No.:
-
基因名:
-
別名:Histone H3.1 (Histone H3/a) (Histone H3/b) (Histone H3/c) (Histone H3/d) (Histone H3/f) (Histone H3/h) (Histone H3/i) (Histone H3/j) (Histone H3/k) (Histone H3/l), HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, HIST1H3J, H3FA, H3FL, H3FC, H3FB, H3FD, H3FI, H3FH, H3FK, H3FF, H3FJ
-
反應種屬:Human
-
免疫原:A synthesized peptide derived from Histone H3 (acetyl K18)
-
免疫原種屬:Homo sapiens (Human)
-
標記方式:Non-conjugated
-
克隆類型:Monoclonal
-
抗體亞型:Rabbit IgG
-
純化方式:Affinity-chromatography
-
克隆號:30A7
-
濃度:It differs from different batches. Please contact us to confirm it.
-
保存緩沖液:Rabbit IgG in 10mM phosphate buffered saline , pH 7.4, 150mM sodium chloride, 0.05% BSA, 0.02% sodium azide and 50% glycerol.
-
產品提供形式:Liquid
-
應用范圍:ELISA, IHC, IF, FC
-
推薦稀釋比:
Application Recommended Dilution IHC 1:50-1:200 IF 1:50-1:200 FC 1:50-1:200 -
Protocols:
-
儲存條件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
貨期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
-
用途:For Research Use Only. Not for use in diagnostic or therapeutic procedures.
相關產品
靶點詳情
-
功能:Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.
-
基因功能參考文獻:
- Data indicate the mechanism for epigenetic regulation in cancer by inducing E3 ubiquitin ligase NEDD4-dependent histone H3 ubiquitination. PMID: 28300060
- the identification of increased expression of H3K27me3 during a patient's clinical course can be helpful for determining whether the tumors are heterochronous PMID: 29482987
- Here, we report that JMJD5, a Jumonji C (JmjC) domain-containing protein, is a Cathepsin L-type protease that mediates histone H3 N-tail proteolytic cleavage under stress conditions that cause a DNA damage response. PMID: 28982940
- Data suggest that Ki-67 antigen proliferative index has important limitations and hhosphohistone H3 (PHH3) is an alternative proliferative marker. PMID: 29040195
- These results identify cytokine-induced histone 3 lysine 27 trimethylation as a mechanism that stabilizes gene silencing in macrophages PMID: 27653678
- This data indicates that, in the early developing human brain, HIST1H3B constitutes the largest proportion of H3.1 transcripts among H3.1 isoforms. PMID: 27251074
- This series of 47 diffuse midline gliomas, histone H3-K27M mutation was mutually exclusive with IDH1-R132H mutation and EGFR amplification, rarely co-occurred with BRAF-V600E mutation, and was commonly associated with p53 overexpression, ATRX loss, and monosomy 10. Among these K27M+ diffuse midline gliomas. PMID: 26517431
- Data show that histone chaperone HIRA co-localizes with viral genomes, binds to incoming viral and deposits histone H3.3 onto these. PMID: 28981850
- These experiments showed that PHF13 binds specifically to DNA and to two types of histone H3 methyl tags (lysine 4-tri-methyl or lysine 4-di-methyl) where it functions as a transcriptional co-regulator. PMID: 27223324
- Hemi-methylated CpGs DNA recognition activates UHRF1 ubiquitylation towards multiple lysines on the H3 tail adjacent to the UHRF1 histone-binding site. PMID: 27595565
- We describe, for the first time, the MR imaging features of pediatric diffuse midline gliomas with histone H3 K27M mutation PMID: 28183840
- Approximately 30% of pediatric high grade gliomas (pedHGG) including GBM and DIPG harbor a lysine 27 mutation (K27M) in histone 3.3 (H3.3) which is correlated with poor outcome and was shown to influence EZH2 function. PMID: 27135271
- H3F3A K27M mutation in adult cerebellar HGG is not rare. PMID: 28547652
- Data show that lysyl oxidase-like 2 (LOXL2) is a histone modifier enzyme that removes trimethylated lysine 4 (K4) in histone H3 (H3K4me3) through an amino-oxidase reaction. PMID: 27735137
- Histone H3 lysine 9 (H3K9) acetylation was most prevalent when the Dbf4 transcription level was highest whereas the H3K9me3 level was greatest during and just after replication. PMID: 27341472
- SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing. PMID: 27614073
- Data suggest that binding of helical tail of histone 3 (H3) with PHD ('plant homeodomain') fingers of BAZ2A or BAZ2B (bromodomain adjacent to zinc finger domain 2A or 2B) requires molecular recognition of secondary structure motifs within H3 tail and could represent an additional layer of regulation in epigenetic processes. PMID: 28341809
- The results demonstrate a novel mechanism by which Kdm4d regulates DNA replication by reducing the H3K9me3 level to facilitate formation of preinitiation complex. PMID: 27679476
- Histone H3 modifications caused by traffic-derived airborne particulate matter exposures in leukocytes PMID: 27918982
- A key role of persistent histone H3 serine 10 or serine 28 phosphorylation in chemical carcinogenesis through regulating gene transcription of DNA damage response genes PMID: 27996159
- hTERT promoter mutations are frequent in medulloblastoma and are associated with older patients, prone to recurrence and located in the right cerebellar hemisphere. On the other hand, histone 3 mutations do not seem to be present in medulloblastoma. PMID: 27694758
- AS1eRNA-driven DNA looping and activating histone modifications promote the expression of DHRS4-AS1 to economically control the DHRS4 gene cluster. PMID: 26864944
- Data suggest that nuclear antigen Sp100C is a multifaceted histone H3 methylation and phosphorylation sensor. PMID: 27129259
- The authors propose that histone H3 threonine 118 phosphorylation via Aurora-A alters the chromatin structure during specific phases of mitosis to promote timely condensin I and cohesin disassociation, which is essential for effective chromosome segregation. PMID: 26878753
- Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its H3 histone recognition. PMID: 27045799
- Functional importance of H3K9me3 in hypoxia, apoptosis and repression of APAK. PMID: 25961932
- Taken together, the authors verified that histone H3 is a real substrate for GzmA in vivo in the Raji cells treated by staurosporin. PMID: 26032366
- We conclude that circulating H3 levels correlate with mortality in sepsis patients and inversely correlate with antithrombin levels and platelet counts. PMID: 26232351
- Data show that double mutations on the residues in the interface (L325A/D328A) decreases the histone H3 H3K4me2/3 demethylation activity of lysine (K)-specific demethylase 5B (KDM5B). PMID: 24952722
- Data indicate that minichromosome maintenance protein 2 (MCM2) binding is not required for incorporation of histone H3.1-H4 into chromatin but is important for stability of H3.1-H4. PMID: 26167883
- Data suggest that histone H3 lysine methylation (H3K4me3) serves a crucial mechanistic role in leukemia stem cell (LSC) maintenance PMID: 26190263
- PIP5K1A modulates ribosomal RNA gene silencing through its interaction with histone H3 lysine 9 trimethylation and heterochromatin protein HP1-alpha. PMID: 26157143
- Data indicate that the lower-resolution mass spectrometry instruments can be utilized for histone post-translational modifications (PTMs) analysis. PMID: 25325711
- Data indicate that inhibition of lysine-specific demethylase 1 activity prevented IL-1beta-induced histone H3 lysine 9 (H3K9) demethylation at microsomal prostaglandin E synthase 1 (mPGES-1) promoter. PMID: 24886859
- The authors report that de novo CENP-A assembly and kinetochore formation on human centromeric alphoid DNA arrays is regulated by a histone H3K9 acetyl/methyl balance. PMID: 22473132
顯示更多
收起更多
-
相關疾病:Glioma (GLM)
-
亞細胞定位:Nucleus. Chromosome.
-
蛋白家族:Histone H3 family
-
數據庫鏈接:
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-