-
中文名稱:SUP45兔多克隆抗體
-
貨號:CSB-PA007840XA01SVG
-
規格:¥440
-
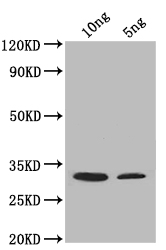
圖片:
-
其他:
產品詳情
-
Uniprot No.:
-
基因名:SUP45
-
別名:Eukaryotic peptide chain release factor subunit 1 (Eukaryotic release factor 1) (eRF1) (Omnipotent suppressor protein 1) SUP45 SAL4 SUP1 YBR143C YBR1120
-
反應種屬:Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
-
免疫原:Recombinant Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) SUP45 protein (26-262aa)
-
免疫原種屬:Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
-
標記方式:Non-conjugated
-
克隆類型:Polyclonal
-
抗體亞型:IgG
-
純化方式:Affinity-chromatography
-
濃度:It differs from different batches. Please contact us to confirm it.
-
保存緩沖液:Preservative: 0.03% Proclin 300
Constituents: 50% Glycerol, 0.01M PBS, pH 7.4 -
產品提供形式:Liquid
-
應用范圍:ELISA, WB
-
Protocols:
-
儲存條件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
貨期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
-
用途:For Research Use Only. Not for use in diagnostic or therapeutic procedures.
相關產品
靶點詳情
-
功能:Directs the termination of nascent peptide synthesis (translation) in response to the termination codons UAA, UAG and UGA.
-
基因功能參考文獻:
- We demonstrated that interaction of [SWI+] and [PIN+] causes inactivation of SUP45 gene that leads to nonsense suppression. Our data show that prion interactions may cause heritable traits in Saccharomyces cerevisiae. PMID: 28027291
- In this work, the authors show that nonsense and missense mutations in the SUP45, but not the SUP35, gene abolish diploid pseudohyphal and haploid invasive growth. PMID: 26054854
- Analysis of the L123 systematic mutants of eRF1 suggested that this amino acid functions in stop codon discrimination in a manner coupled with eRF3 binding, and distinctive from previously reported adjacent residues. PMID: 25897120
- We characterized a region of the eRF1 N-terminal domain, the P1 pocket, that we had previously shown to be involved in termination efficiency. We identified two residues, arginine 65 and lysine 109, as critical for recognition of the three stop codons. PMID: 25735746
- Prion-like determinant [NSI+] decreases the relative amounts of SUP45 mRNA that causes a decrease in the amounts of Sup45 protein. PMID: 25842864
- HLJ1 and TEF2 genes are affecting Sup35 protein prionization and/or the efficiency of translation termination. PMID: 24466750
- A deterministic mathematical model of the eRF1 (SUP45) feedback loop was developed using a staged increase in model complexity. PMID: 23104998
- The results indicated that changes near cavities two and three frequently mediated significant effects on stop codon selectivity by eRF1 (SUP45). Changes in the YCF motif correlated most consistently with variant code stop codon selectivity. PMID: 22543865
- Deletion of each UPF gene leads to allosuppresson of ade1-14 mutation without changing eRF1 amount PMID: 22670525
- SUP45 gene modulates nonsense suppression in S. cerevisiae PMID: 22215010
- The role of translation termination factor eRF1 in the regulation of pseudohyphal growth in Saccharomyces cerevisiae cells. PMID: 20714858
- Results indicate that point mutants of Saccharomyces cerevisiae eRF1 display significant variability in their stop codon read-through phenotypes. PMID: 20444877
- Overexpression of genes encoding tRNA(Tyr) AND tRNA(Gln) improves viability of nonsense mutants in SUP45 gene in yeast Saccharomyces cerevisiae PMID: 20586191
- Mtq2p methylates Sup45p PMID: 16321977
- sup45 mutations do not only change translation fidelity but also acts by causing a change in mRNA stability PMID: 17705828
- The results of this study suggest that the N-terminal eRF1 fragment is indispensable for cell viability of nonsense mutants due to the involvement in termination of translation. PMID: 18069340
- The data suggest that increasing stop codon suppression and decreasing of the binding affinity of eRF1(Y410S) were probably due to the substitution of tyrosine to serine which triggered the structural change on the C-terminal domain PMID: 18463713
- mutagenesis of SUP45 led to the identification of two new pockets in domain 1 (P1 and P2) involved in the regulation of eRF1 activity. PMID: 19174561
- compared readthrough efficiency of the natural termination codon of SUP45 gene, spontaneous sup45-n (nonsense) mutations, nonsense mutations obtained by site-directed mutagenesis (76Q --> TAA, 242R --> TGA, 317L --> TAG) PMID: 19370360
顯示更多
收起更多
-
亞細胞定位:Cytoplasm.
-
蛋白家族:Eukaryotic release factor 1 family
-
數據庫鏈接:
KEGG: sce:YBR143C
STRING: 4932.YBR143C
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC, IF, FC
Species Reactivity: Human, Mouse, Rat
-
-
-
-
-
-