SUMO2 Antibody
-
中文名稱:SUMO2兔多克隆抗體
-
貨號:CSB-PA099258
-
規格:¥2024
-
圖片:
-
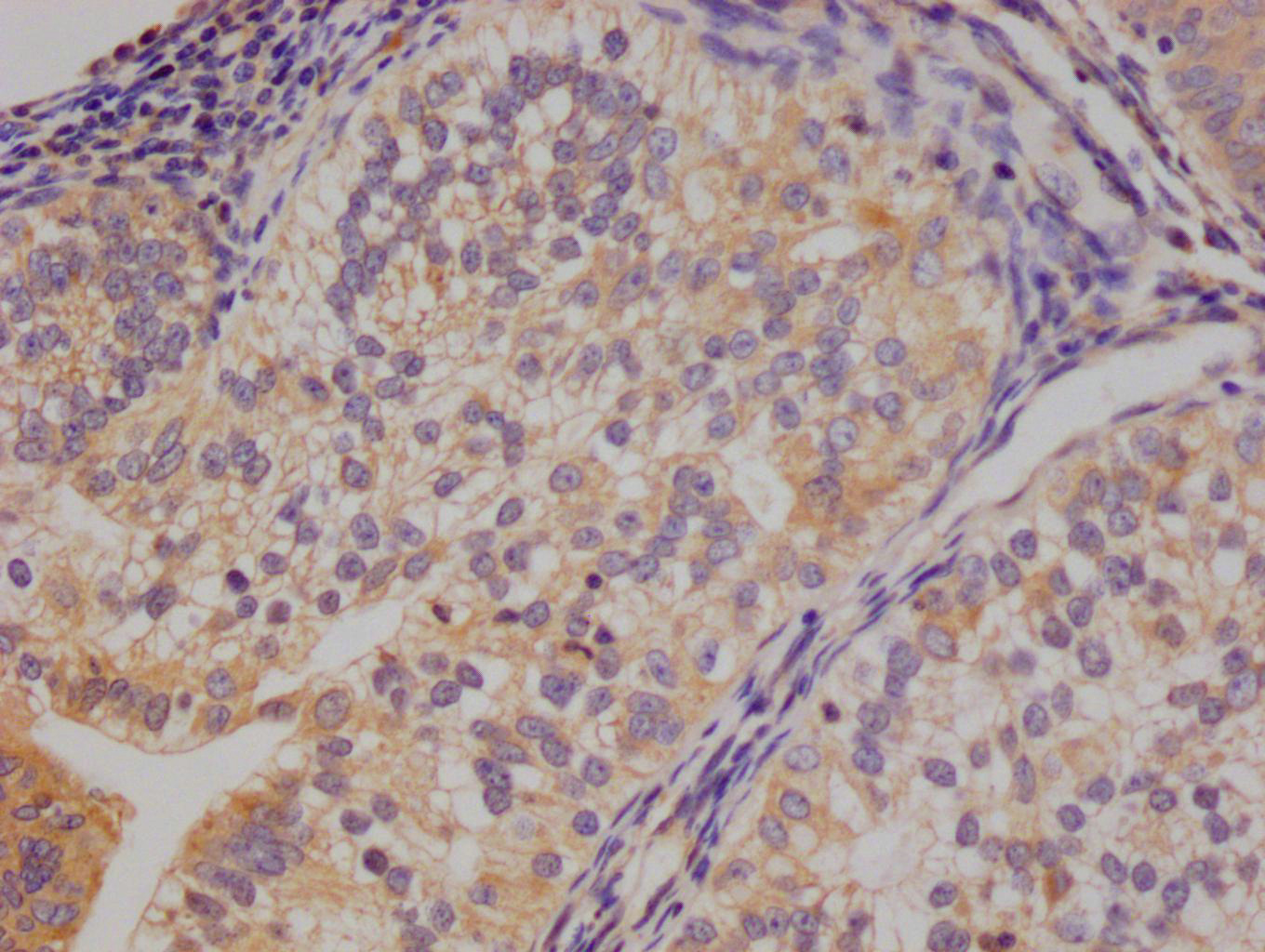
Gel: 15%+12%SDS-PAGE Lysates (from left to right): Human small intestine stromal cancer tissue Amount of lysate: 40μg per lane Primary antibody: 1/800 dilution Secondary antibody dilution: 1/8000 Exposure time: 10 seconds
-
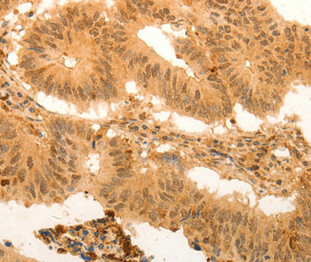
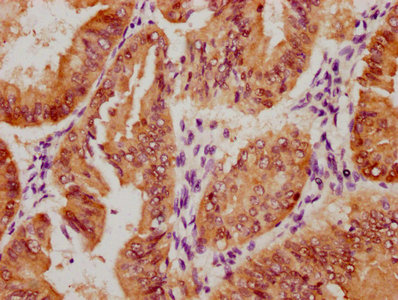
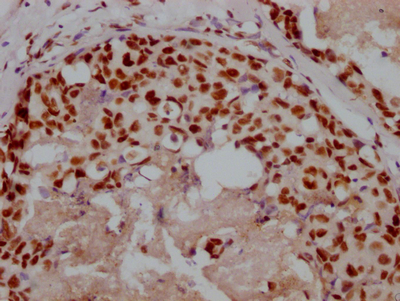
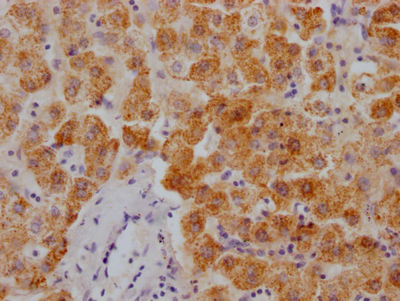
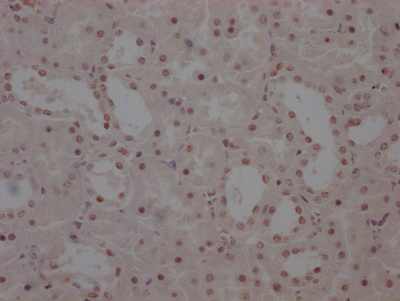
Immunohistochemical analysis of paraffin-embedded Human colon cancer tissue using at dilution 1/100.
-
-
其他:
產品詳情
-
產品名稱:Rabbit anti-Homo sapiens (Human) SUMO2 Polyclonal antibody
-
Uniprot No.:
-
基因名:SUMO2
-
宿主:Rabbit
-
反應種屬:Human,Mouse,Rat
-
免疫原:Synthetic peptide corresponding to a region derived from internal residues of Human SMT3 suppressor of mif two 3 homolog 2/3/4 (S. cerevisiae)
-
免疫原種屬:Homo sapiens (Human)
-
克隆類型:Polyclonal
-
純化方式:Antigen Affinity Purified
-
濃度:It differs from different batches. Please contact us to confirm it.
-
產品提供形式:Liquid
-
應用范圍:ELISA,WB,IHC
-
推薦稀釋比:
Application Recommended Dilution WB 1:500-1:3000 IHC 1:50-1:100 -
Protocols:
-
儲存條件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
貨期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
-
用途:For Research Use Only. Not for use in diagnostic or therapeutic procedures.
相關產品
靶點詳情
-
功能:Ubiquitin-like protein that can be covalently attached to proteins as a monomer or as a lysine-linked polymer. Covalent attachment via an isopeptide bond to its substrates requires prior activation by the E1 complex SAE1-SAE2 and linkage to the E2 enzyme UBE2I, and can be promoted by an E3 ligase such as PIAS1-4, RANBP2, CBX4 or ZNF451. This post-translational modification on lysine residues of proteins plays a crucial role in a number of cellular processes such as nuclear transport, DNA replication and repair, mitosis and signal transduction. Polymeric SUMO2 chains are also susceptible to polyubiquitination which functions as a signal for proteasomal degradation of modified proteins. Plays a role in the regulation of sumoylation status of SETX.
-
基因功能參考文獻:
- It has been shown that conjugation of SUMO2, but not SUMO1 or SUMO3, to the essential replication factor PCNA is induced on transcribed chromatin by the RNAPII-bound helicase RECQ5. PMID: 30006506
- Identified SUMO2 as a most efficient calcineurin-NFAT (Cn-NFAT) activator. SUMO2-mediated activation of Cn-NFAT signaling in cardiomyocytes translated into a hypertrophic phenotype. PMID: 27767176
- We further show that the SUMO2 pathway acts independently of exogenous C-MYC expression and in parallel with small-molecule enhancers of reprogramming. Importantly, suppression of SUMO2 also promotes the generation of human induced pluripotent stem cells . PMID: 26947976
- Sumoylation of PML with SUMO2 by UBC9/UBE2I can lead to formation of polymeric SUMO chains. Data suggest that coordination of growing poly-SUMO chain with "back side" binding site on UBC9/UBE2I appears to be required for SUMO chain elongation on PML. (PML = promyelocytic leukemia protein; SUMO2 = small ubiquitin-like modifier 2; UBC9/UBE2I = ubiquitin-conjugating enzyme UBC9/UBE2I) PMID: 28784659
- The data demonstrate that SUMO2 conjugation and SENP3-driven deSUMOylation of PELP1 is instrumental for ordered progression of ribosome maturation, and they provide molecular insight into the dynamics of ribosome maturation. PMID: 27814492
- This study reveals an essential role of SUMOylated FADD in Drp1- and caspase-10-dependent necrosis. PMID: 27799292
- This study demonstrated that two polymorphisms of SUMO2 were significantly associated with an increased risk of AD in female group. PMID: 27084229
- Hsp27-Ubc9 pathway recognizes the conformation of mutant CFTR which leads to its SUMO-2 conjugation and degradation by the ubiquitin-proteasome system. PMID: 26627832
- Data suggest that RAP80 SIM (SUMO interacting motif) binds SUMO-2; both specificity and affinity are enhanced through phosphorylation of canonical CK2 (casein kinase 2) site within the SIM. PMID: 26719330
- Data show that substantial increases in the binding of small ubiquitin-like modifier 2 (SUMO-2) to active DNA regulatory elements in response to heat stress. PMID: 26152697
- Data suggest that small ubiquitin-related modifier protein SUMO1 modification of the promyelocytic leukemia protein (PML) RING domain promotes SUMO2 conjugation to Lys160. PMID: 26060329
- present a comprehensive proteomic analysis of changes in the cellular SUMO2 proteome during HSV-1 infection PMID: 26200910
- Genome-wide analysis of sumoylation dynamics in response to replication stress reveals novel SUMO-2 modified target proteins and acceptor lysines relevant for genome stability. PMID: 25755297
- Kif18A is covalently modified by SUMO2 during the cell cycle, and its sumoylation peaks at metaphase and then rapidly decreases upon anaphase onset PMID: 25884224
- SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. PMID: 25722289
- SUMO2 target regulating STAT3 signaling will contribute to the comprehension of SUMO2 function in T cells, particularly in Tc17 cell development and antitumor activity. PMID: 25762490
- These results confirm that the SUMO machinery is involved in TRIM5alpha-mediated retroviral restriction, and demonstrate that TRIM5alpha is a SUMO 1 and SUMO 2 substrate. PMID: 25880753
- SUMO2 is robustly conjugated to p35 in HEK293 cells. Sumoylation is a likely candidate mechanism for the rapid modulation of p35/Cdk5 activity in physiological situations as well as in disease. PMID: 25391294
- Studied chromosome spreads to more precisely define the localization of SUMO-2/3 (small ubiquitin-related modifier) to the inner centromere and protein scaffold of mitotic chromosomes. PMID: 25367092
- SUMO-2 promotes mRNA translation by enhancing interaction between eIF4E and eIF4G PMID: 24971752
- 22 SUMO2 targets with increased SUMOylation during DNA replication stress were identified, many of which play key functions within the DNA replication machinery and/or in the cellular response to DNA damage. PMID: 25497329
- The SUMO2 N-terminal tail does not affect significantly the secondary and tertiary structure of the globular domain, neither its conformational stability nor its function, but it contributes to decrease aggregation tendency without compromising function. PMID: 24564702
- Using mice haploinsufficient for the SUMO E2 enzyme, we found that sumoylation regulates intestinal permeability and is required to restrict epithelial invasion and control mucosal inflammation PMID: 25097252
- Data indicate the identification of 1,002 small ubiquitin-like modifier 2 protein SUMO2T90K modified sites in cells. PMID: 24782567
- the structure of SENP2-Loop1 in complex with SUMO2 was solved at 2.15 A resolution, and reveals the details of an interface exclusive to SENP6/7 and the formation of unique contacts between both proteins PMID: 24424631
- identified 295 SUMO1 and 167 SUMO2 sites on endogenous substrates of HeLa cells PMID: 25114211
- Modification of TDG by small ubiquitin-like modifier (SUMO) proteins weakens its binding to abasic DNA. PMID: 24753249
- Proteomic and co-immunoprecipitation analysis further reveal that the SUMO-2 modified transcription repressor KAP1 is a critical factor recruited by LANA(SIM). PMID: 24278015
- FoxM1 is a key mitotic SUMO2 target protein. PMID: 24582501
- PIP5K1A is modified by polySUMO-2 only during apoptosis. PMID: 23994136
- lysine 91, the major target of Nurr1 SUMOylation is contained in a canonical synergy control motif, indicating that SUMO-2 posttranslational modification of Nurr1 regulates its transcriptional synergy in complex promoters PMID: 23358114
- Co-immunoprecipitations and in vitro SUMOylation confirmed ARHGAP21 specific modification by SUMO2/3 and mapped the SUMOylation site to ARHGAP21 lysine K1443. PMID: 22922005
- IRF8 is conjugated to SUMO2/3 in resting macrophages. PMID: 22942423
- SUMO2 binding by the Epstein-Barr virus protein kinase BGLF4 is crucial for BGLF4 function. PMID: 22398289
- TRIM28 acts as a SUMO E3 ligase by increasing SUMOylation of IRF7 both in vivo and in vitro, with little effect on the closely related IRF3. PMID: 21940674
- The authors demonstrate that LANA2 is covalently conjugated to SUMO1 and SUMO2 both in vitro and in latently KSHV-infected B-cells. PMID: 20881090
- Report a mass spectrometry method to indentify SUMO-2 acceptor lysines in endogenous proteins and reveal an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. PMID: 20797634
- HSF1 is modified by SUMO-1 and SUMO-2 in a stress-inducible manner. PMID: 12665592
- CENPC target sites that can be sumoylated by SUMO-2 were shown to be equally susceptible to SUMO-1 attachments which include specific sites on SUMO-2 itself, Ubc9, and the recombinant CENP-C fragments PMID: 15272016
- Through a comprehensive structure-function analysis, we have identified a single critical sector along the second beta sheet and the following alpha helix of SUMO2 PMID: 15870296
- Using yeast two-hybrid system, bioinformatics, and NMR spectroscopy we define a common SUMO-interacting motif (SIM) and map its binding surfaces on SUMO1 and SUMO2 PMID: 16524884
- x-ray crystallographic structure of SENP1-SUMO-2 complex demonstrates structural basis for discrimination between SUMO paralogues during processing PMID: 16553580
- SUMO-2-associated proteins identified in this study may contribute to SUMO-dependent regulation of transcription or other processes PMID: 16567619
- Myeloid elf-1-like factor (MEF) or Elf4 is modified by conjugation with SUMO-1/-2 (small ubiquitin-related modifier). PMID: 16904644
- Results support the notion that SUMO-2 are evolutionarily designed for function both structurally and thermodynamically in their low-populated, high-energy conformers rather than in their basic folded conformers. PMID: 18081309
- Nm23-H1 was modified with SUMO-2 after X-ray irradiation. PMID: 19332021
- Direct interactions between CoREST1 and SUMO-2 mediate SUMO-dependent changes in chromatin structure and transcription that are important for cell-type-specific gene expression. PMID: 19394292
- A massive redistribution of SUMO-2 was observed that affected many biological pathways that are important for the heat shock response. PMID: 19638612
顯示更多
收起更多
-
亞細胞定位:Nucleus. Nucleus, PML body.
-
蛋白家族:Ubiquitin family, SUMO subfamily
-
組織特異性:Broadly expressed.
-
數據庫鏈接:
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-